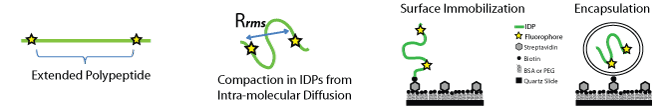
Intrinsically Disordered Proteins (IDPs)
Classical studies of enzyme activity lead to the understanding that biological polypeptide chains fold into a stable 3D structure based on repeating elements of secondary structure. A precise tertiary structure is needed to position key atoms for molecular recognition and catalysis. The enzyme and substrate fit together like the proverbial lock and key. This model has proven able to describe the majority of known proteins, but it is now recognized that not all proteins adopt a stable 3D structure. Such Intrinsically disordered polypeptides (IDPs) are constantly sampling alternate conformations, giving rise to a dynamic “supertertiary” structure . Disorder does not mean that a protein is broken. If the structure determines a protein’s function, then adopting different structures would allow different functions from the same protein. Disorder was a late evolutionary acquisition that gave a selective advantage.
In the Bowen Lab
Our research uses hybrid methods, meaning that we combine data from different kinds of experiments, to study the structure and dynamics of disordered proteins. We use the tools and data from classical structural biology to refine molecular models based on data from single-molecule fluorescence microscopy. We are investigating the how disordered regions contribute to the allosteric regulation of protein activity, and how posttranslational modifications alter the supertertiary structure of disordered proteins. Understanding how intrinsic disorder in proteins supports complex signaling in the brain will facilitate the discovery of novel approaches to the treatment of neurological, psychiatric and neurodegenerative disorders.
Structural Ensembles
IDPs play critical roles in molecular recognition during neurotransmitter signaling. . Localization depends on interaction with molecular scaffold proteins like PSD-95. PSD-95 contains structured binding domains connected by strings of disorder, which permit a flexible organization of scaffolding sites. The supertertiary arrangment of domains can be regulated by postranslational modifications and may place steric restrictions on which protein complexes are favored.
Allosteric Regulation
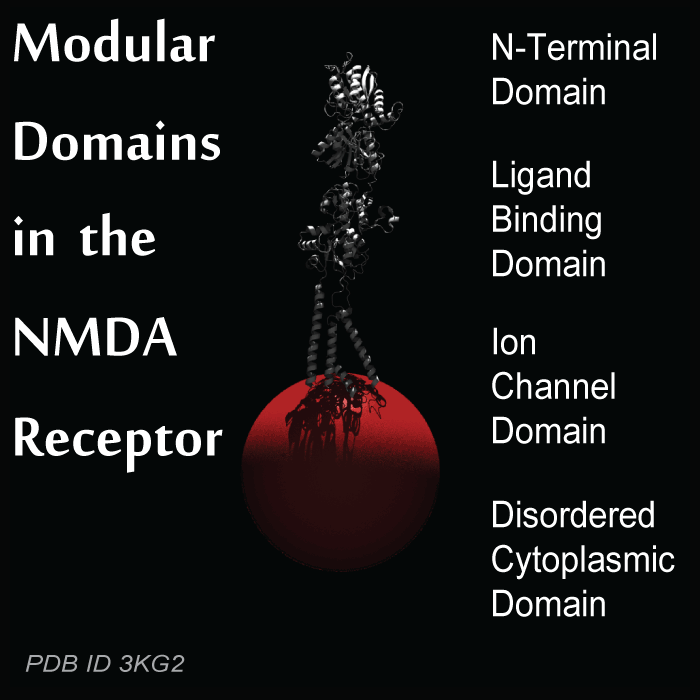
NMDA-sensitive Ionotropic glutamate receptors (NMDARs) are central to mediating the plasticity that underlies higher brain functions like learning and memory and development of neural circuits. Some NMDARs contain large disordered regions that regulate receptor activity and localization in the brain. Many regulatory domains are disordered because intrinsic disorder provides allosteric coupling between domains. Thus, each NMDAR can adopt a different conformation and the conformations can change in response to activity.